Top 10 Publikationen
Ma-Lauer Y, Li P, Niemeyer D, Richter A, Pusl K, von Brunn B, Ru Y, Xiang C, Schwinghammer S, Liu J, Baral P, Berthold EJ, Qiu H, Roy A, Kremmer E, Flaswinkel H, Drosten C, Jin Z, von Brunn A. (2024). Oxysterole-binding protein targeted by SARS-CoV-2 viral proteins regulates coronavirus replication. Front. Cell. Infect. Microbiol. 2024; 14:1383917. https://doi.org/10.3389/fcimb.2024.1383917
Berthold EJ, Ma-Lauer Y, Chakraborty A, von Brunn B, Hilgendorff A, Hatz R, Behr J, Hausch F, Staab-Weijnitz CA*, von Brunn A* (2022). Effects of immunophilin inhibitors and nonimmunosuppressive analogs on coronavirus replication in human infection models. Front. Cell. Infect. Microbiol. 2022, 12:958634. https://doi.org/10.3389/fcimb.2022.958634
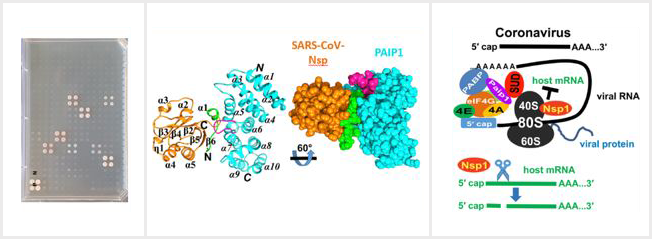
Lei J†, Ma-Lauer Y†, Han Y, Thoms M, Buschauer R, Jores J, Thiel V, Beckmann R, Deng W, Leonhardt H, Hilgenfeld R*, von Brunn A* (2021). The SARS-unique domain (SUD) of SARS-CoV and SARS-CoV-2 interacts with human PAIP1 to enhance viral RNA translation. The EMBO Journal 1;40(11):e102277. https://doi.org/10.15252/embj.2019102277
Poulsen, NN, von Brunn, A, Hornum, M and Blomberg, JM (2020) Cyclosporine and COVID-19: Risk or Favorable? Am J Transplant. (2020). 20:2975-2982. https://doi.org/10.1111/ajt.16250
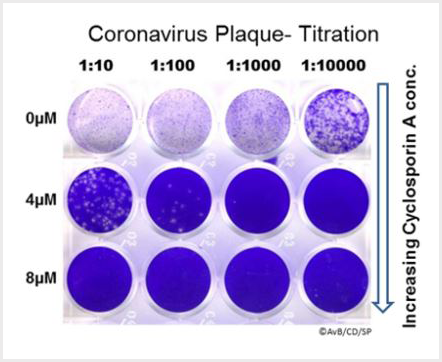
Ma-Lauer Y, Zheng, Y Malešević M, von Brunn B, Fischer F, von Brunn A* (2020). Influences of cyclosporin A and non-immunosuppressive derivatives on cellular cyclophilins and viral nucleocapsid protein during human coronavirus 229E replication. Antiviral Research 173, 104620. https://doi.org/10.1016/j.antiviral.2019.104620
Ma-Lauer Y, Carbajo-Lozoya J, Hein M, Müller MA, Deng W, Lei J, Meyer B, Kusov Y, von Brunn B, Bairad DR, Hünten S, Drosten C, Hermeking H, Leonhardt, H, Mann M, Hilgenfeld R, von Brunn, A* (2016). p53 down-regulates SARS-Coronavirus replication and is targeted by the SARS-Unique Domain and PLpro via E3 ubiquitin ligase RCHY1 Proc Natl Acad Sci U S A, 113(35): E5192-201. www.pnas.org/cgi/doi/10.1073/pnas.1603435113
Carbajo-Lozoya, J., Ma-Lauer, Y., Malešević, M., Theuerkorn, M., Kahlert, V., Prell, E., von Brunn, B., Muth, D., Baumert, T.F., Drosten, C., Fischer, G., and von Brunn, A* (2014). Human coronavirus NL63 replication is cyclophilin A-dependent and inhibited by non-immunosuppressive cyclosporine A-derivatives including Alisporivir. Virus Research 184, 44-53. https://doi.org/10.1016/j.virusres.2014.02.010
Carbajo-Lozoya, J., Müller, M.A., Kallies, S., Thiel, V., Drosten, C. and von Brunn, A* (2012). Replication of human coronaviruses SARS-CoV, HCoV-NL63 and HCoV-229E is inhibited by the drug FK506. Virus Research 165, 112–117. https://doi.org/10.1016/j.virusres.2012.02.002
Pfefferle S, Schöpf J, Kögl M, Friedel C, Müller MA, Carbajo-Lozoya J, Stellberger T, von Dall'Armi E, Herzog P, Kallies S, Niemeyer D, Ditt V, Kuri T, Züst R, Pumpor K, Hilgenfeld R, Schwarz F, Zimmer, R., Steffen I, Weber F, Thiel V, Herrler G, Thiel H-J, Schwegmann-Weßels, C., Pöhlmann S, Haas J, Drosten C, von Brunn, A* (2011). The SARS-Coronavirus-Host Interactome: Identification of Cyclophilins as Target for Pan-Coronavirus Inhibitors. PLoS Pathogens 7(10): e1002331. https://doi.org/10.1371/journal.ppat.1002331
von Brunn A*, Teepe C, Simpson JC, Pepperkok R, Friedel C., Zimmer R, Roberts R, Baric R, Haas, J* (2007). Analysis of Intraviral Protein-Protein Interactions of the SARS Coronavirus ORFeome. PLoS ONE 23;2:e459. https://doi.org/10.1371/journal.pone.0000459